News
Understanding the patterns of spread of influenza viruses – Insights from Dr David Collins
October 21, 2020
Dr David Collins Owour is a virologist interested in the applications of advanced genomics and bioinformatics to investigate emerging and re-emerging viral pathogens associated with infectious diseases. He holds a bachelor’s degree in Biomedical Technology and a masters degree in Molecular Genetics and Diagnostics. His current research seeks to: (i) determine how closely “related” or similar viruses are to one another genetically; (ii) monitor how viruses change over time; and (iii) monitor for genetic changes in zoonotic viruses circulating in animal populations that could enable them infect humans. He successfully defended his PhD with just minor corrections which focussed on Viral Genomics and Bioinformatics. It sought to understand how human influenza viruses genetically change over time and how they spread at different levels of transmission.
The research career journey
My interest in infectious diseases developed back in the 1990s, at a time when the HIV/Aids epidemic was at its peak in Kenya. I was concerned by the horrifying reports about the deadly epidemic and other diseases of major concern in Kenya and globally. Fascinated by the informative posters on controlling the disease and impressed by the work done by healthcare professionals fighting the disease, I was motivated to pursue a career in science, with the hope of joining the healthcare profession after my training.
The problem
Influenza is among the leading causes of respiratory illness and death globally, despite the availability of influenza vaccines and antiviral drugs. Globally, annual influenza epidemics result in 3 to 5 million cases of severe illness, and 290,000 to 650,000 respiratory deaths. The analysis of influenza virus genomes and associated epidemiological data is important to better understand the geographical source and patterns of spread of influenza. For example, understanding the geographical source of new influenza viruses could potentially improve the early detection of new viruses and enable prediction of vaccine strains for use in future vaccines. However, due to insufficient influenza virus genome data from understudied tropical and sub-tropical regions, for example, sub-Saharan Africa, relatively little is known about the possible role the regions play in the global spread of influenza.
The research utilized influenza virus genome data from Kenya and Africa (among the first-ever influenza virus genomes generated within Kenya) to improve our understanding of the global patterns of spread of influenza viruses.
Understanding the spread of influenza virus
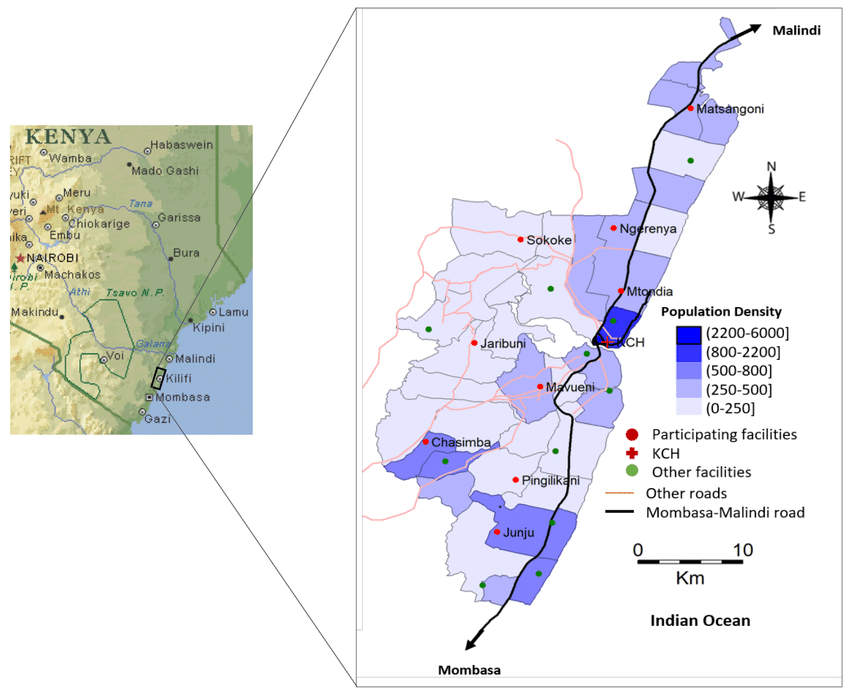
The research sought to understand the patterns of spread of influenza viruses at multiple levels of transmission: (i) at a local community, for example, in a rural community setting in Kilifi, coastal Kenya; (ii) countrywide in Kenya; (iii) continentwide in Africa; and (iv) globally across all continents and to show their interconnectedness. In the local community, influenza epidemics are initiated by multiple virus introductions, which then spread to multiple locations within the community in a short period of time and usually from populous to less populous locations. Countrywide, influenza spreads from multiple locations to multiple destinations within Kenya and between locations in proximity, the year-round virus spread regulated by virus introductions from outside Kenya. Within Africa, influenza viruses spread extensively across the continent with significant transmission from the northern to the southern hemisphere countries. Globally, influenza viruses spread from multiple geographical regions to multiple geographical destinations that include Africa, which suggests that multiple geographical regions, including Africa, are important drivers behind the global spread of influenza viruses.
The role of Africa and other understudied regions in the global spread of influenza is not well understood due to insufficient virus genome data despite the high disease burden of influenza. Until this work, the role of Africa in the global spread of influenza was poorly understood due to insufficient data. The findings from this study support the notion that multiple geographical regions, including Africa, are important for driving the global spread of influenza viruses whereby new virus strains can emerge in any geographical region. Therefore, there is need for improved surveillance throughout Africa, especially in sub-Saharan Africa, and other understudied regions to (i) improve early detection of new virus strains that may arise in these regions; and (ii) improve the selection of the most effective vaccine strains for circulating seasonal influenza viruses, which are currently based on vaccine strains selected from countries in the temperate northern and southern hemispheres.
Why training could change the future of the continent
Postdoctoral training is an opportunity to further the PhD research work, create research collaborations with other institutions, and to apply for independent funding to support research. However, postdoctoral training, like PhD training, is limited in Africa. Therefore, additional funding is required to fund world-class PhD studies, which can then be advanced into world-class postdoctoral research that attract independent funding.
More support from African governments needed
More often than not, the African PhD student is older and with more societal obligations compared to their counterparts in other regions, thus a need for more funding to support their work. While the efforts of initiatives like IDeAL that support African research are commendable, it is time for African governments to offer more support to research. The ongoing COVID-19 pandemic is a wakeup call for the need for additional support; Africa needs a critical mass of world-class scientists to tackle its healthcare challenges, which will be achieved through research support.
- 51
- 66
- 48
- 104
- 44
- 423


Share: